Myeloma Genomics and cell signaling
Poster Session 3
P-375: Unraveling Signaling Pathways in Multiple Myeloma Cardiovascular Toxicities from Pharmacovigilance and Pharmacogenomics Data Mining
Friday, September 29, 2023
1:15 PM - 2:15 PM EEST
- XX
Xuan Xu, PhD
Postdoctoral Associate
Kansas State University
Olathe, Kansas, United States
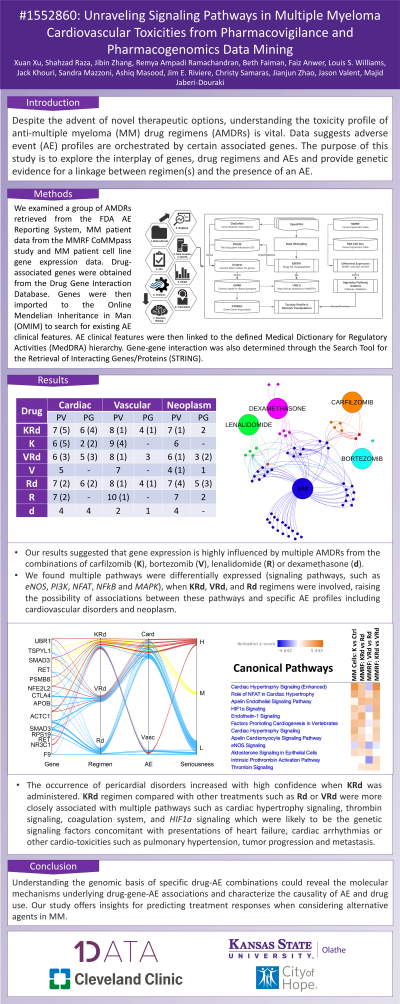
Introduction: Despite the advent of novel therapeutic options, understanding the toxicity profile of anti-multiple myeloma (MM) drug regimens (AMDRs) is vital. Data suggests adverse event (AE) profiles are orchestrated by certain associated genes. The purpose of this study is to explore the interplay of genes, drug regimens and AEs and provide genetic evidence for a linkage between regimen(s) and the presence of an AE.
Methods: We examined a group of AMDRs retrieved from the FDA AE Reporting System, MM patient data from the MMRF CoMMpass study and MM patient cell line gene expression data. Drug-associated genes were obtained from the Drug Gene Interaction Database. Genes were then imported to the Online Mendelian Inheritance in Man (OMIM) to search for existing AE clinical features. AE clinical features were then linked to the defined Medical Dictionary for Regulatory Activities (MedDRA) hierarchy. Gene-gene interaction was also determined through the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING).
Results: Our results suggested that gene expression is highly influenced by multiple AMDRs from the combinations of carfilzomib, bortezomib, lenalidomide or dexamethasone. We found multiple pathways were differentially expressed (signaling pathways, such as eNOS, PI3K, NFAT, NFkB and MAPK), when carfilzomib-lenalidomide-dexamethasone, bortezomib-lenalidomide-dexamethasone, and lenalidomide-dexamethasone regimens were involved, raising the possibility of associations between these pathways and specific AE profiles including cardiovascular disorders, neoplasm, or other serious AEs, see Table for pharmacovigilance (PV) and pharmacogenomics (PG). The occurrence of pericardial disorders increased with high confidence when carfilzomib-lenalidomide-dexamethasone was administered. We also found that the carfilzomib-lenalidomide-dexamethasone regimen compared with other treatments such as lenalidomide-dexamethasone or bortezomib-lenalidomide-dexamethasone were more closely associated with multiple pathways such as cardiac hypertrophy signaling, thrombin signaling, coagulation system, and HIF1a signaling which were likely to be the genetic signaling factors concomitant with presentations of heart failure, cardiac arrhythmias or other cardio-toxicities such as pulmonary hypertension, tumor progression and metastasis.
Drug Cardiac Vascular Neoplasm
PV PG PV PG PV PG
Carfilzomib-Lenalidomide-Dexamethasone 7 (5) 6 (4) 8 (1) 4 (1) 7 (1) 2
Carfilzomib 6 (5) 2 (2) 9 (4) - 6 -
Bortezomib-Lenalidomide-Dexamethasone 6 (3) 5 (3) 8 (1) 3 6 (1) 3 (2)
Bortezomib 5 - 7 - 4 (1) 1
Lenalidomide-Dexamethasone 7 (2) 6 (2) 8 (1) 4 (1) 7 (4) 5 (3)
Lenalidomide 7 (2) - 10 (1) - 7 2
Dexamethasone 4 4 2 1 4 -
Conclusions: Understanding the genomic basis of specific drug-AE combinations could reveal the molecular mechanisms underlying drug-gene-AE associations and characterize the causality of AE and drug use. Our study offers insights for predicting treatment responses when considering alternative agents in MM.
Methods: We examined a group of AMDRs retrieved from the FDA AE Reporting System, MM patient data from the MMRF CoMMpass study and MM patient cell line gene expression data. Drug-associated genes were obtained from the Drug Gene Interaction Database. Genes were then imported to the Online Mendelian Inheritance in Man (OMIM) to search for existing AE clinical features. AE clinical features were then linked to the defined Medical Dictionary for Regulatory Activities (MedDRA) hierarchy. Gene-gene interaction was also determined through the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING).
Results: Our results suggested that gene expression is highly influenced by multiple AMDRs from the combinations of carfilzomib, bortezomib, lenalidomide or dexamethasone. We found multiple pathways were differentially expressed (signaling pathways, such as eNOS, PI3K, NFAT, NFkB and MAPK), when carfilzomib-lenalidomide-dexamethasone, bortezomib-lenalidomide-dexamethasone, and lenalidomide-dexamethasone regimens were involved, raising the possibility of associations between these pathways and specific AE profiles including cardiovascular disorders, neoplasm, or other serious AEs, see Table for pharmacovigilance (PV) and pharmacogenomics (PG). The occurrence of pericardial disorders increased with high confidence when carfilzomib-lenalidomide-dexamethasone was administered. We also found that the carfilzomib-lenalidomide-dexamethasone regimen compared with other treatments such as lenalidomide-dexamethasone or bortezomib-lenalidomide-dexamethasone were more closely associated with multiple pathways such as cardiac hypertrophy signaling, thrombin signaling, coagulation system, and HIF1a signaling which were likely to be the genetic signaling factors concomitant with presentations of heart failure, cardiac arrhythmias or other cardio-toxicities such as pulmonary hypertension, tumor progression and metastasis.
Drug Cardiac Vascular Neoplasm
PV PG PV PG PV PG
Carfilzomib-Lenalidomide-Dexamethasone 7 (5) 6 (4) 8 (1) 4 (1) 7 (1) 2
Carfilzomib 6 (5) 2 (2) 9 (4) - 6 -
Bortezomib-Lenalidomide-Dexamethasone 6 (3) 5 (3) 8 (1) 3 6 (1) 3 (2)
Bortezomib 5 - 7 - 4 (1) 1
Lenalidomide-Dexamethasone 7 (2) 6 (2) 8 (1) 4 (1) 7 (4) 5 (3)
Lenalidomide 7 (2) - 10 (1) - 7 2
Dexamethasone 4 4 2 1 4 -
Conclusions: Understanding the genomic basis of specific drug-AE combinations could reveal the molecular mechanisms underlying drug-gene-AE associations and characterize the causality of AE and drug use. Our study offers insights for predicting treatment responses when considering alternative agents in MM.
